NPs Basic Information
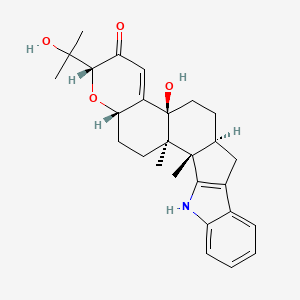
|
Name |
Paxilline
|
| Molecular Formula | C27H33NO4 | |
| IUPAC Name* |
(1S,2R,5S,7R,11S,14S)-11-hydroxy-7-(2-hydroxypropan-2-yl)-1,2-dimethyl-6-oxa-23-azahexacyclo[12.10.0.02,11.05,10.016,24.017,22]tetracosa-9,16(24),17,19,21-pentaen-8-one
|
|
| SMILES |
C[C@]12CC[C@H]3C(=CC(=O)[C@H](O3)C(C)(C)O)[C@@]1(CC[C@@H]4[C@@]2(C5=C(C4)C6=CC=CC=C6N5)C)O
|
|
| InChI |
InChI=1S/C27H33NO4/c1-24(2,30)23-20(29)14-18-21(32-23)10-11-25(3)26(4)15(9-12-27(18,25)31)13-17-16-7-5-6-8-19(16)28-22(17)26/h5-8,14-15,21,23,28,30-31H,9-13H2,1-4H3/t15-,21-,23-,25+,26+,27+/m0/s1
|
|
| InChIKey |
ACNHBCIZLNNLRS-UBGQALKQSA-N
|
|
| Synonyms |
paxilline; 57186-25-1; NSC 658707; BRN 5317894; MLS000028812; C27H33NO4; CHEBI:34907; 3T9U9Z96L7; (1s,2r,5s,7r,11s,14s)-11-hydroxy-7-(2-hydroxypropan-2-yl)-1,2-dimethyl-6-oxa-23-azahexacyclo[12.10.0.02,11.05,10.016,24.017,22]tetracosa-9,16(24),17,19,21-pentaen-8-one; 1233509-81-3; SMR000058863; paxillin; 2H-Pyrano[2'',3'':5',6']benz[1',2':6,7]indeno[1,2-b]indol-3(4bH)-one, 5,6,6a,7,12,12b,12c,13,14,14a-decahydro-4b-hydroxy-2-(1-hydroxy-1-methylethyl)-12b,12c-dimethyl-, (2R,4bS,6aS,12bS,12cR,14aS)-rel-; PAXILINE; UNII-3T9U9Z96L7; NSC-658707; (+)-paxilline; 2H-Pyrano(2'',3'':5',6')benz(1',2':6,7)indeno(1,2-b)indol-3(4bH)-one, 5,6,6a,7,12,12b,12c,13,14,14a-decahydro-4b-hydroxy-2-(1-hydroxy-1-methylethyl)-12b,12c-dimethyl-, (2R,4bS,6aS,12bS,12cR,14aS)-; MFCD00083464; Spectrum5_001975; CBiol_001842; BSPBio_001427; KBioGR_000147; KBioSS_000147; SCHEMBL361232; CHEMBL410063; cid_105008; BDBM50854; KBio2_000147; KBio2_002715; KBio2_005283; KBio3_000293; KBio3_000294; DTXSID10972643; Bio1_000128; Bio1_000617; Bio1_001106; Bio2_000147; Bio2_000627; HMS1361H09; HMS1791H09; HMS1989H09; HMS2232J18; HMS3268F05; HMS3402H09; HY-N6778; ZINC3996035; HB1056; KC-155; AKOS024456907; Paxilline, powder, >=98% (HPLC); IDI1_033897; NCGC00025342-05; NCGC00025342-07; 2H-1-Benzopyrano(5',6':6,7)indeno(1,2-b)indol-3(4bh)-one, 5,6,6a,7,12,12b,12c,13,14,14a-decahydro-4b-hydroxy-2-(1-hydroxy-1-methylethyl)-12b,12c-dimethyl-, (2-alpha,4b-beta,6a-alpha,12b-beta,12c-alpha,14a-beta)-; CS-0026740; SR-01000597527; SR-01000597527-1; BRD-K38251852-001-02-5; BRD-K38251852-001-06-6; Q10860377; EDD20523-48C0-4594-B22A-1FE297F60611; (2R,4bS,6aS,12bS,12cR,14aS)-4b-hydroxy-2-(2-hydroxypropan-2-yl)-12b,12c-dimethyl-5,6,6a,7,12,12b,12c,13,14,14a-decahydro-2H-[1]benzopyrano[5',6':6,7]indeno[1,2-b]indol-3(4bH)-one; (2R,4bS,6aS,12bS,12cR,14aS)-4b-hydroxy-2-(2-hydroxypropan-2-yl)-12b,12c-dimethyl-5,6,6a,7,12,12b,12c,13,14,14a-decahydro-2H-chromeno[5',6':6,7]indeno[1,2-b]indol-3(4bH)-one; (2R,4bS,6aS,12bS,12cR,14aS)-5,6,6a, 7,12,12b,12c,13,14,14a-Decahydro-4b-hydroxy-2-(1-h ydroxy-1-methylethyl)-12b,12c-dimethyl-2H-pyrano[2 '',3'':5',6']benz[1',2':6,7]indeno[1,2-b]indol-3(4bH)-one; (2R,4bS,6aS,12bS,12cR,14aS)-5,6,6a,7,12,12b,12c,13,14,14a-Decahydro-4b-hydroxy-2-(1-hydroxy-1-methylethyl)-12b,12c-dimethyl-2H-pyrano[2'',3'':5',6']benz[1',2':6,7]indeno[1,2-b]indol-3(4bH)-one; NCGC00025342-07_C27H33NO4_(2R,4bS,6aS,12bS,12cR,14aS)-4b-Hydroxy-2-(2-hydroxy-2-propanyl)-12b,12c-dimethyl-5,6,6a,7,12,12b,12c,13,14,14a-decahydro-2H-chromeno[5',6':6,7]indeno[1,2-b]indol-3(4bH)-one
|
|
| CAS | 57186-25-1 | |
| PubChem CID | 105008 | |
| ChEMBL ID | CHEMBL410063 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 435.6 | ALogp: | 3.5 |
| HBD: | 3 | HBA: | 4 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 82.6 | Aromatic Rings: | 6 |
| Heavy Atoms: | 32 | QED Weighted: | 0.612 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.882 | MDCK Permeability: | 0.00002600 |
| Pgp-inhibitor: | 0.232 | Pgp-substrate: | 0.934 |
| Human Intestinal Absorption (HIA): | 0.008 | 20% Bioavailability (F20%): | 0.003 |
| 30% Bioavailability (F30%): | 0.288 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.954 | Plasma Protein Binding (PPB): | 78.78% |
| Volume Distribution (VD): | 1.1 | Fu: | 9.12% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.153 | CYP1A2-substrate: | 0.883 |
| CYP2C19-inhibitor: | 0.486 | CYP2C19-substrate: | 0.807 |
| CYP2C9-inhibitor: | 0.544 | CYP2C9-substrate: | 0.248 |
| CYP2D6-inhibitor: | 0.069 | CYP2D6-substrate: | 0.108 |
| CYP3A4-inhibitor: | 0.901 | CYP3A4-substrate: | 0.92 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 7.922 | Half-life (T1/2): | 0.426 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.038 | Human Hepatotoxicity (H-HT): | 0.29 |
| Drug-inuced Liver Injury (DILI): | 0.398 | AMES Toxicity: | 0.508 |
| Rat Oral Acute Toxicity: | 0.96 | Maximum Recommended Daily Dose: | 0.937 |
| Skin Sensitization: | 0.81 | Carcinogencity: | 0.897 |
| Eye Corrosion: | 0.005 | Eye Irritation: | 0.013 |
| Respiratory Toxicity: | 0.988 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001492 | 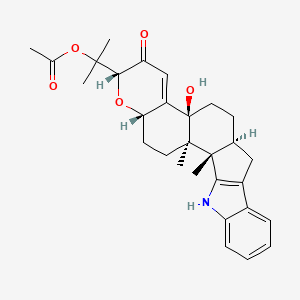 |
0.824 | D0H4JM | 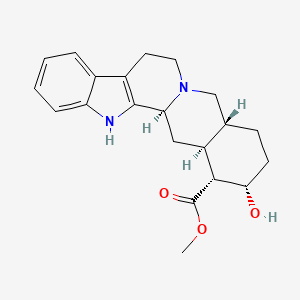 |
0.281 | ||
| ENC005990 | 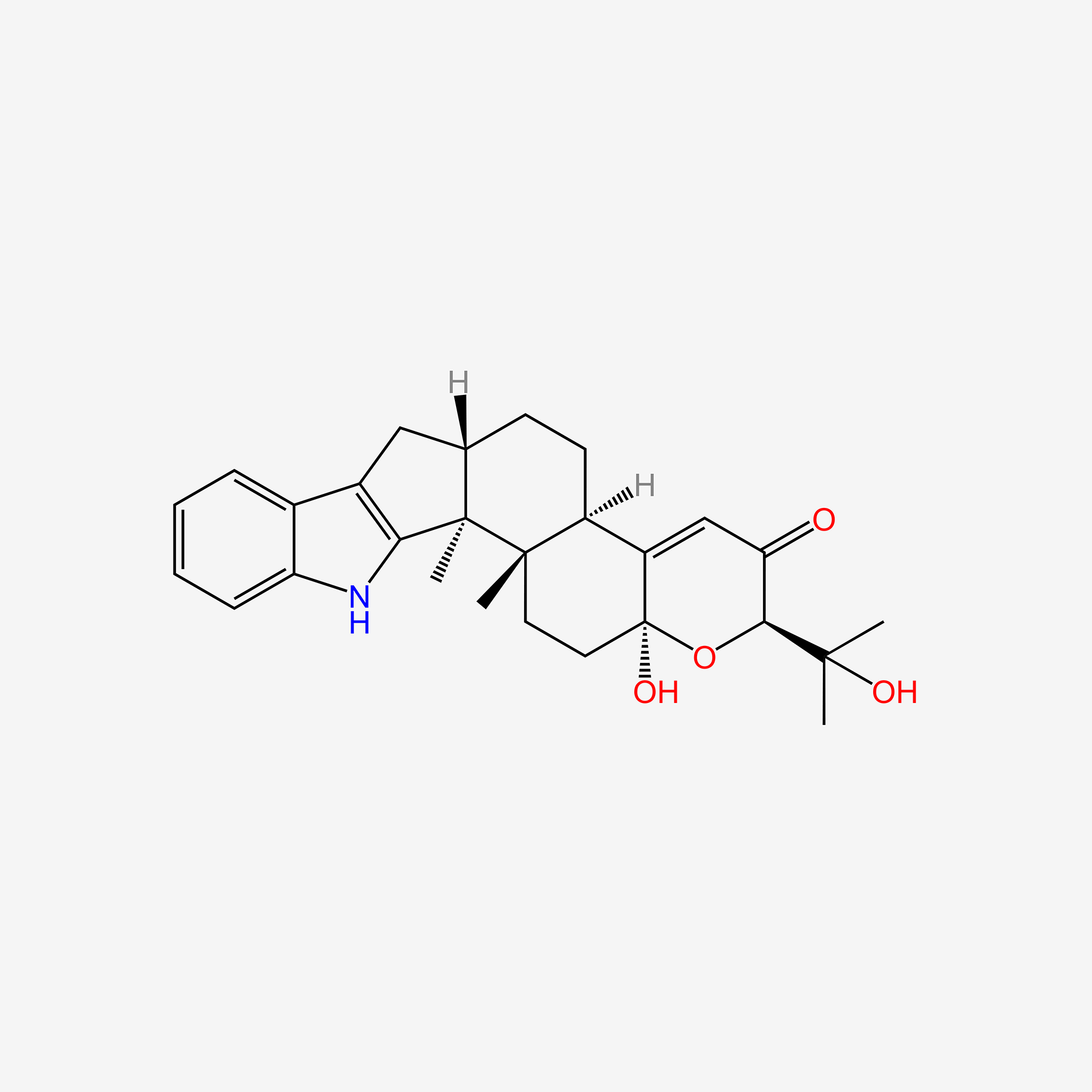 |
0.712 | D0U7GP | 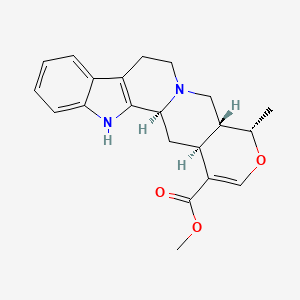 |
0.281 | ||
| ENC002279 | 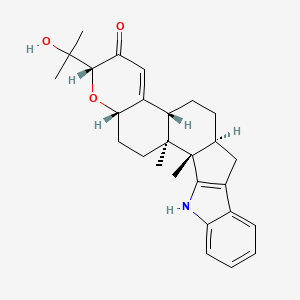 |
0.709 | D01JGV | 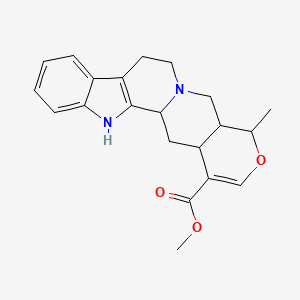 |
0.281 | ||
| ENC005988 | 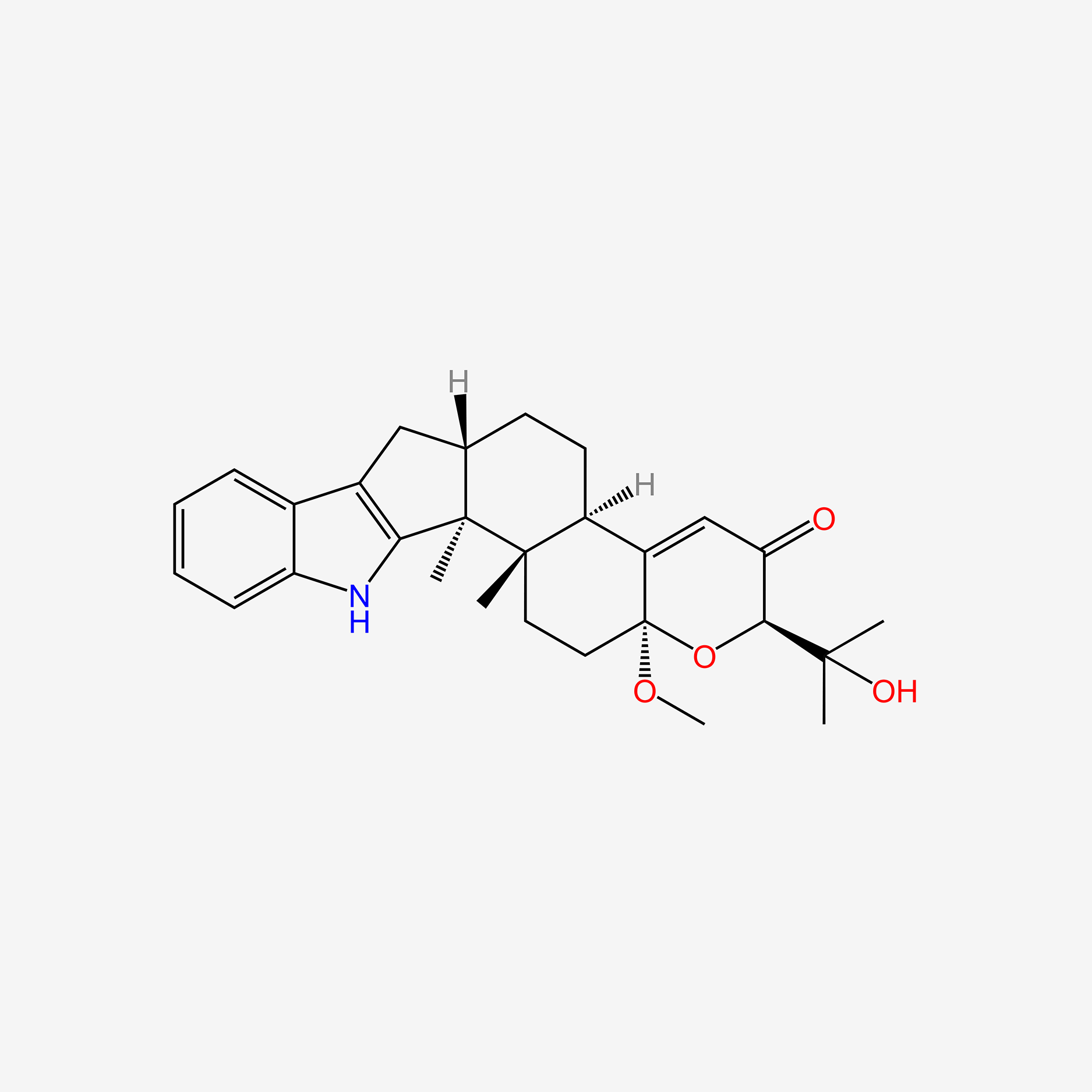 |
0.661 | D05MQK | 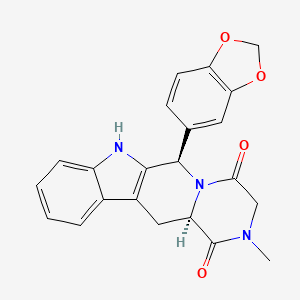 |
0.263 | ||
| ENC001966 | 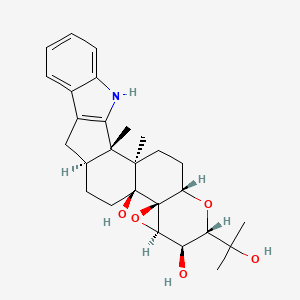 |
0.588 | D0V4WD | 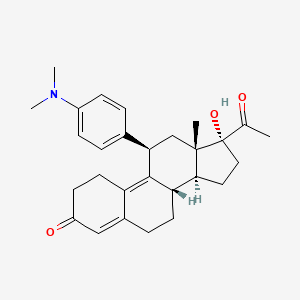 |
0.254 | ||
| ENC005989 | 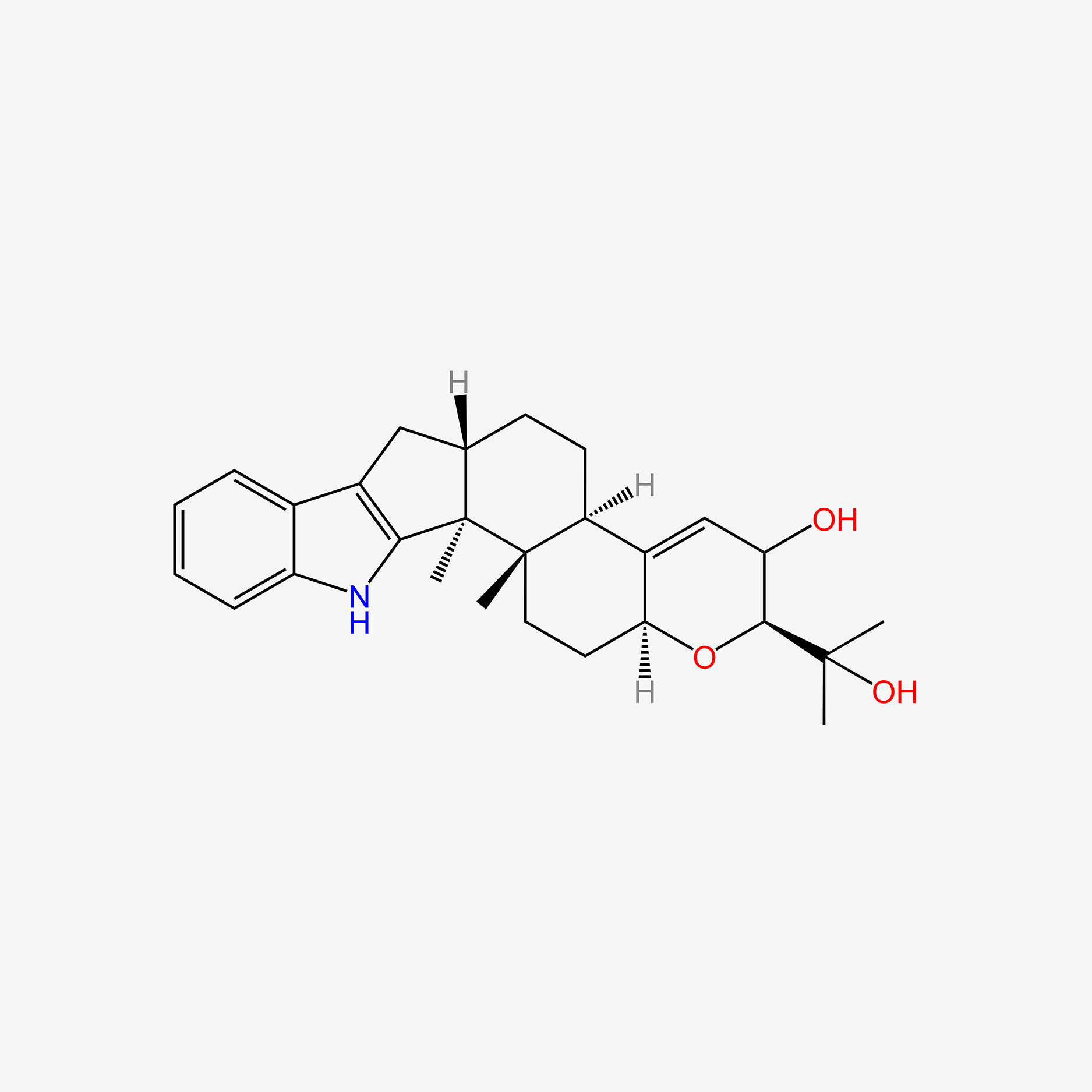 |
0.571 | D0K0KH | 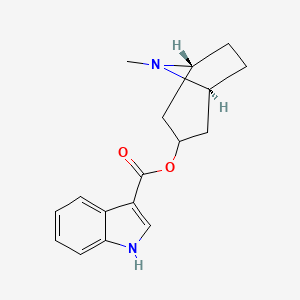 |
0.250 | ||
| ENC003172 | 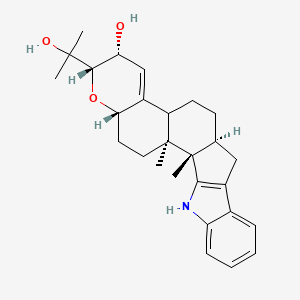 |
0.571 | D04RLY | 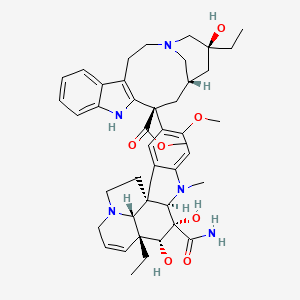 |
0.246 | ||
| ENC002951 | 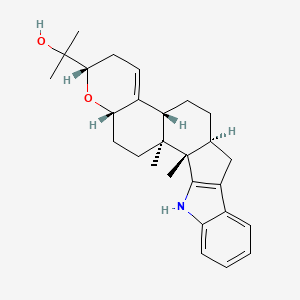 |
0.568 | D04GJN | 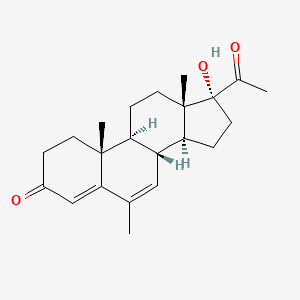 |
0.246 | ||
| ENC000857 | 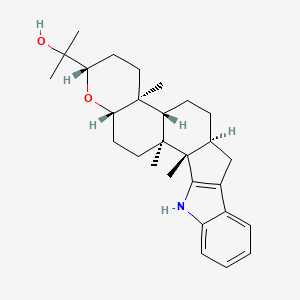 |
0.530 | D06AEO | 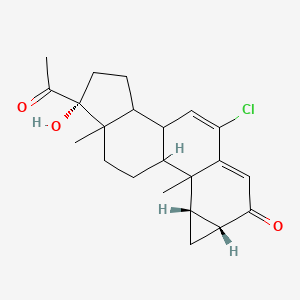 |
0.240 | ||
| ENC004710 | 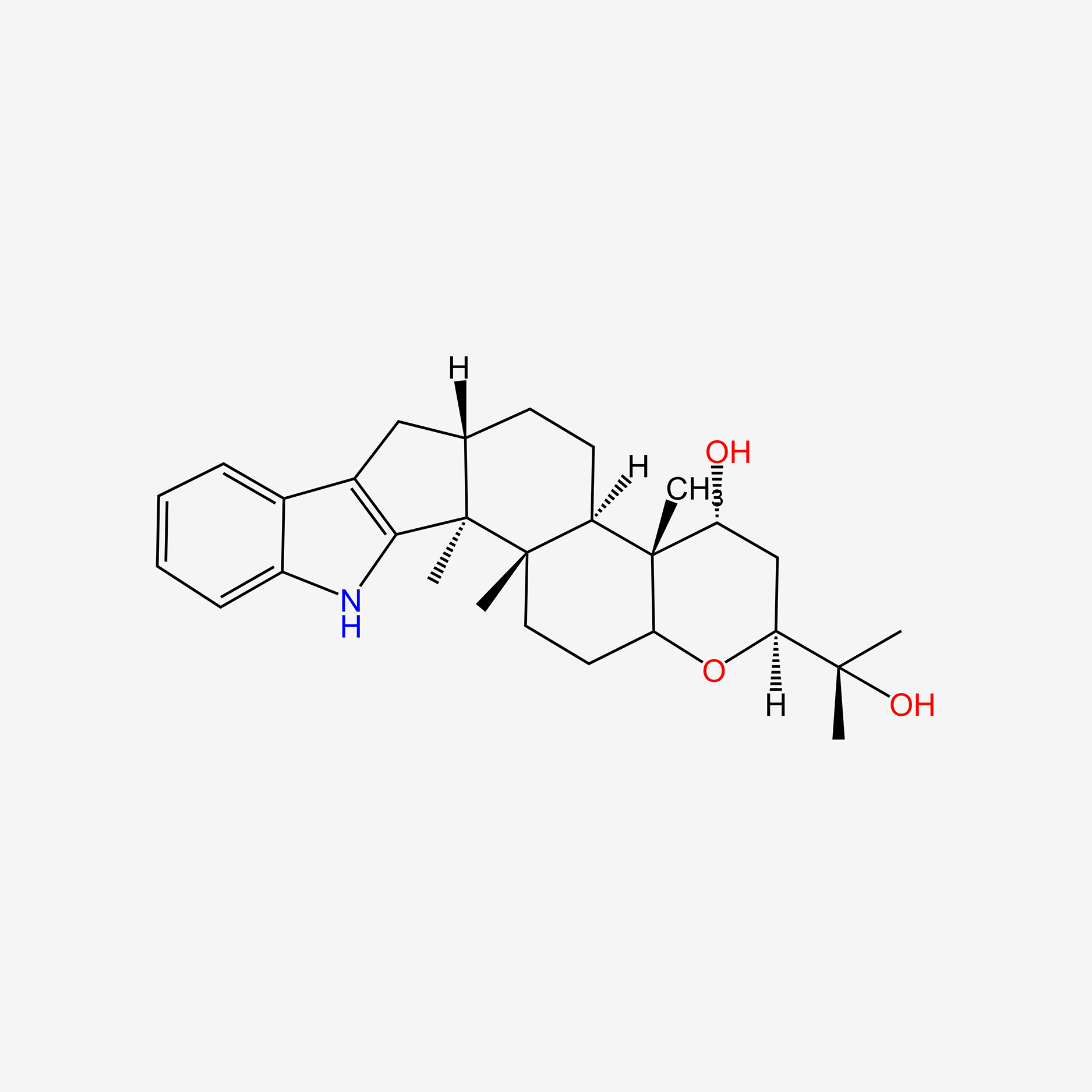 |
0.521 | D0OT9S |  |
0.240 | ||