NPs Basic Information
|
Name |
Hexyl isovalerate
|
| Molecular Formula | C11H22O2 | |
| IUPAC Name* |
hexyl 3-methylbutanoate
|
|
| SMILES |
CCCCCCOC(=O)CC(C)C
|
|
| InChI |
InChI=1S/C11H22O2/c1-4-5-6-7-8-13-11(12)9-10(2)3/h10H,4-9H2,1-3H3
|
|
| InChIKey |
RSDDTPVXLMVLQE-UHFFFAOYSA-N
|
|
| Synonyms |
Hexyl isovalerate; Hexyl 3-methylbutanoate; 10032-13-0; Butanoic acid, 3-methyl-, hexyl ester; Hexyl isopentanoate; n-Hexyl isopentanoate; Isovaleric acid, hexyl ester; 10632-13-0; Hexyl isovaleriate; Hexyl isovalerianate; Hexyl 3-methyl butanoate; 3-Methylbutyric acid hexyl ester; n-Hexyl iso-valerate; FEMA No. 3500; HEXYL ISOVALERATE STANDARD FOR GC; Hexyl 3-methylbutyrate; 5FJ2M7YCY6; Hexyl 3-methylbutanoate (natural); EINECS 233-105-7; UNII-5FJ2M7YCY6; BRN 1759161; AI3-33585; Caproyl isovalerate; 1-Hexanol isovalerate; Hexyl 3-methyl butyrate; SCHEMBL573930; HEXYL ISOVALERATE [FCC]; DTXSID4064921; HEXYL ISOVALERATE [FHFI]; FEMA 3500; 3-Methylbutanoic acid hexyl ester; CHEBI:179380; Hexyl 3-methylbutanoate, >=99%; ZINC1850757; Isovaleric acid, hexyl ester (8CI); LMFA07010908; FT-0627062; FT-0693397; J-000099; Q27261969
|
|
| CAS | 10032-13-0 | |
| PubChem CID | 61455 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 186.29 | ALogp: | 3.7 |
| HBD: | 0 | HBA: | 2 |
| Rotatable Bonds: | 8 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 26.3 | Aromatic Rings: | 0 |
| Heavy Atoms: | 13 | QED Weighted: | 0.444 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.319 | MDCK Permeability: | 0.00002700 |
| Pgp-inhibitor: | 0.04 | Pgp-substrate: | 0.002 |
| Human Intestinal Absorption (HIA): | 0.002 | 20% Bioavailability (F20%): | 0.676 |
| 30% Bioavailability (F30%): | 0.855 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.847 | Plasma Protein Binding (PPB): | 90.52% |
| Volume Distribution (VD): | 0.599 | Fu: | 10.78% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.956 | CYP1A2-substrate: | 0.355 |
| CYP2C19-inhibitor: | 0.788 | CYP2C19-substrate: | 0.483 |
| CYP2C9-inhibitor: | 0.703 | CYP2C9-substrate: | 0.771 |
| CYP2D6-inhibitor: | 0.011 | CYP2D6-substrate: | 0.076 |
| CYP3A4-inhibitor: | 0.096 | CYP3A4-substrate: | 0.187 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 11.967 | Half-life (T1/2): | 0.634 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.055 | Human Hepatotoxicity (H-HT): | 0.028 |
| Drug-inuced Liver Injury (DILI): | 0.333 | AMES Toxicity: | 0.005 |
| Rat Oral Acute Toxicity: | 0.07 | Maximum Recommended Daily Dose: | 0.012 |
| Skin Sensitization: | 0.913 | Carcinogencity: | 0.258 |
| Eye Corrosion: | 0.979 | Eye Irritation: | 0.988 |
| Respiratory Toxicity: | 0.23 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
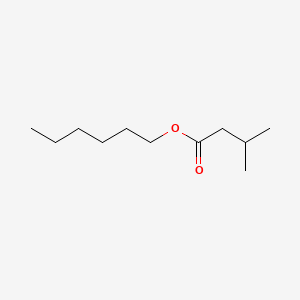
| ENC000570 | 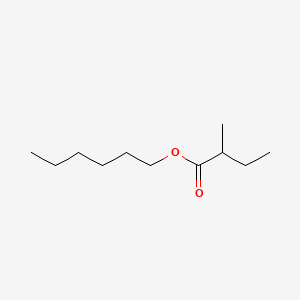 |
0.628 | D0AY9Q | 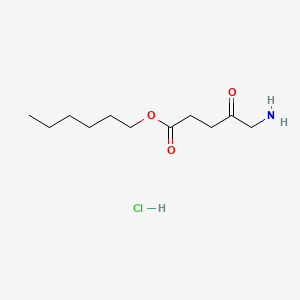 |
0.481 | ||
| ENC000726 | 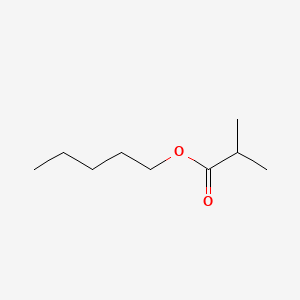 |
0.600 | D01QLH | 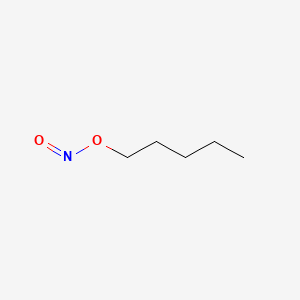 |
0.357 | ||
| ENC000655 | 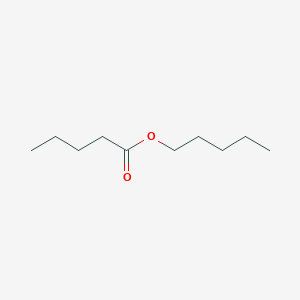 |
0.581 | D05ATI |  |
0.302 | ||
| ENC000228 | 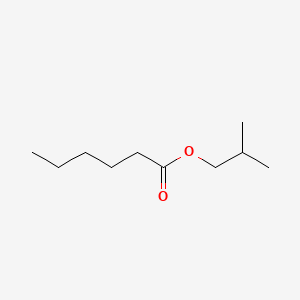 |
0.558 | D06ORU | 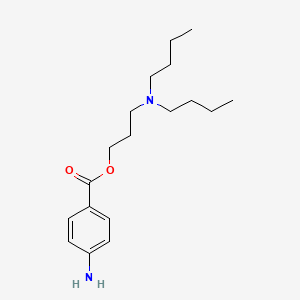 |
0.276 | ||
| ENC000718 | 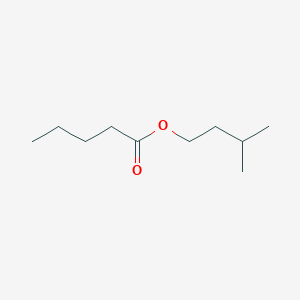 |
0.558 | D0G2KD | 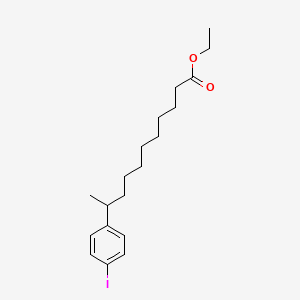 |
0.276 | ||
| ENC000241 | 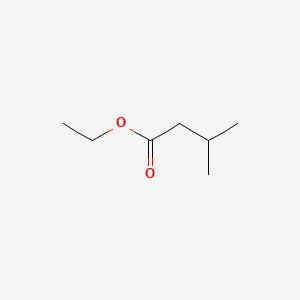 |
0.526 | D0Z5SM |  |
0.271 | ||
| ENC000459 |  |
0.512 | D0H2YX | 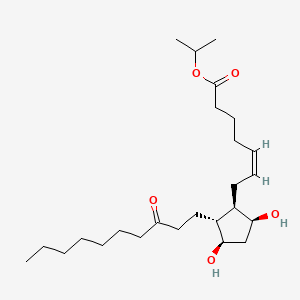 |
0.269 | ||
| ENC000742 | 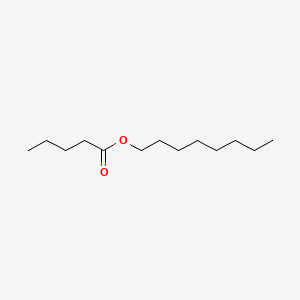 |
0.510 | D0X4FM | 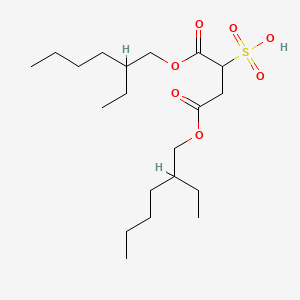 |
0.264 | ||
| ENC000448 | 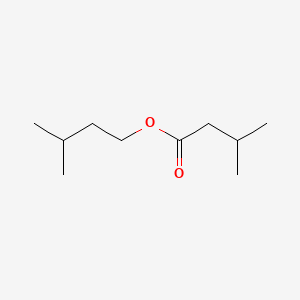 |
0.500 | D05PLH | 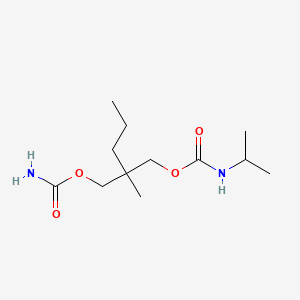 |
0.262 | ||
| ENC000231 | 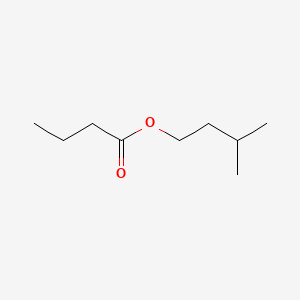 |
0.488 | D00WUF | 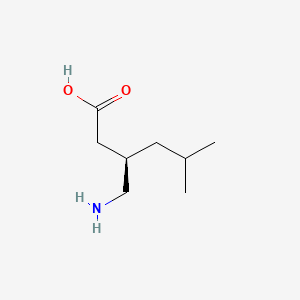 |
0.260 | ||