NPs Basic Information
|
Name |
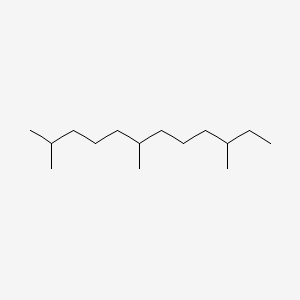
2,6,10-Trimethyldodecane
|
| Molecular Formula | C15H32 | |
| IUPAC Name* |
2,6,10-trimethyldodecane
|
|
| SMILES |
CCC(C)CCCC(C)CCCC(C)C
|
|
| InChI |
InChI=1S/C15H32/c1-6-14(4)10-8-12-15(5)11-7-9-13(2)3/h13-15H,6-12H2,1-5H3
|
|
| InChIKey |
YFHFHLSMISYUAQ-UHFFFAOYSA-N
|
|
| Synonyms |
2,6,10-TRIMETHYLDODECANE; Farnesane; 3891-98-3; Farnesan; Dodecane, 2,6,10-trimethyl-; Dodecane,2,6,10-trimethyl-; 8X81V0IT6Q; HEMISQUALANE; NEOSSANCE TMD; EC 622-542-2; UNII-8X81V0IT6Q; 2,6,10-trimethyl-dodecane; DTXSID1058634; CHEBI:36756; MFCD00027077; AKOS024334439; AS-58771; CS-0452006; FT-0668468; E78326; Q27116951; 7B5
|
|
| CAS | 3891-98-3 | |
| PubChem CID | 19773 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 212.41 | ALogp: | 7.5 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 9 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 0.0 | Aromatic Rings: | 0 |
| Heavy Atoms: | 15 | QED Weighted: | 0.452 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.424 | MDCK Permeability: | 0.00000924 |
| Pgp-inhibitor: | 0.025 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.002 | 20% Bioavailability (F20%): | 0.311 |
| 30% Bioavailability (F30%): | 0.89 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.453 | Plasma Protein Binding (PPB): | 97.76% |
| Volume Distribution (VD): | 2.667 | Fu: | 2.14% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.583 | CYP1A2-substrate: | 0.215 |
| CYP2C19-inhibitor: | 0.447 | CYP2C19-substrate: | 0.754 |
| CYP2C9-inhibitor: | 0.539 | CYP2C9-substrate: | 0.907 |
| CYP2D6-inhibitor: | 0.087 | CYP2D6-substrate: | 0.039 |
| CYP3A4-inhibitor: | 0.148 | CYP3A4-substrate: | 0.148 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 7.198 | Half-life (T1/2): | 0.067 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.028 | Human Hepatotoxicity (H-HT): | 0.019 |
| Drug-inuced Liver Injury (DILI): | 0.124 | AMES Toxicity: | 0.005 |
| Rat Oral Acute Toxicity: | 0.024 | Maximum Recommended Daily Dose: | 0.022 |
| Skin Sensitization: | 0.882 | Carcinogencity: | 0.045 |
| Eye Corrosion: | 0.991 | Eye Irritation: | 0.968 |
| Respiratory Toxicity: | 0.242 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000806 |  |
0.818 | D00FSV |  |
0.379 | ||
| ENC000537 |  |
0.804 | D03LGY | 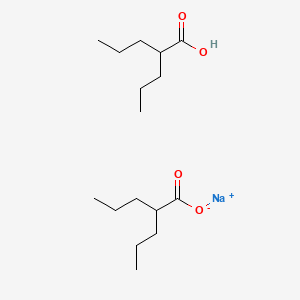 |
0.240 | ||
| ENC000766 | 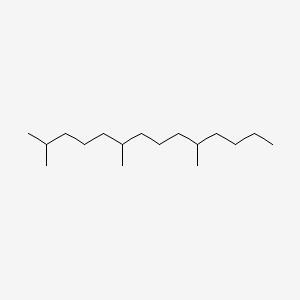 |
0.755 | D0X4FM | 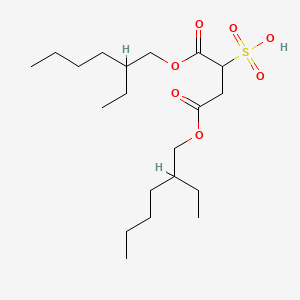 |
0.211 | ||
| ENC000441 | 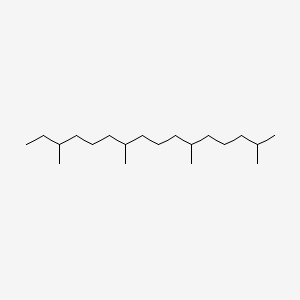 |
0.741 | D0K5WS |  |
0.192 | ||
| ENC000902 | 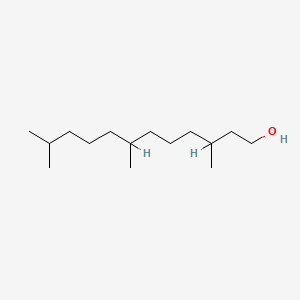 |
0.729 | D0ZI4H | 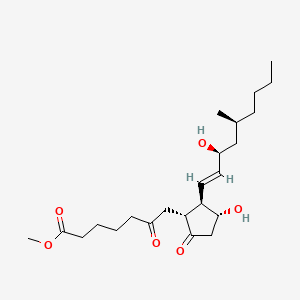 |
0.190 | ||
| ENC000538 | 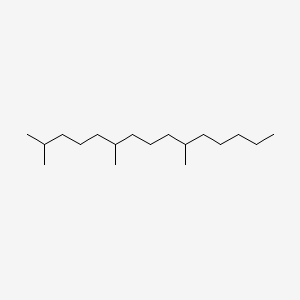 |
0.712 | D05QNO | 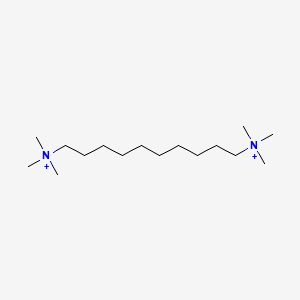 |
0.189 | ||
| ENC000622 |  |
0.702 | D0R6BR |  |
0.188 | ||
| ENC000627 |  |
0.702 | D0N3NO |  |
0.188 | ||
| ENC001286 | 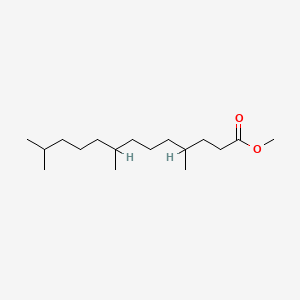 |
0.655 | D0D9NY | 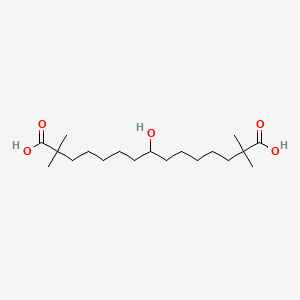 |
0.184 | ||
| ENC000503 | 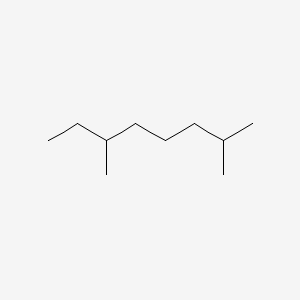 |
0.650 | D0K3ZR | 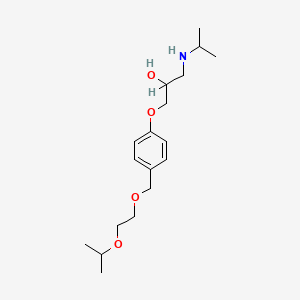 |
0.182 | ||