- 1.1 Search by NPs name
- 1.2 Search by NPs structure
- 1.3 Search by Endophyte
- 1.4 Search by Target
- 1.5 Search by Bioactivity
- 2.1 Browse Endophyte by Taxonomy
- 2.2 Browse NP by Classification
- 2.3 Browse Target Cell Line by Category
- 2.4 Browse Target Protein by Protein Family
- 2.5 Browse Target Organism by Taxonomy
- 3.1 Webpage of a specific endophyte
- 3.2 Webpage of a specific NP
- 3.3 Webpage of a specific target protein
- 3.4 Webpage of a specific target cell line
- 3.5 Webpage of a specific target organism
EMNPD MANUAL FOR USERS
1. SEARCH EMNPD
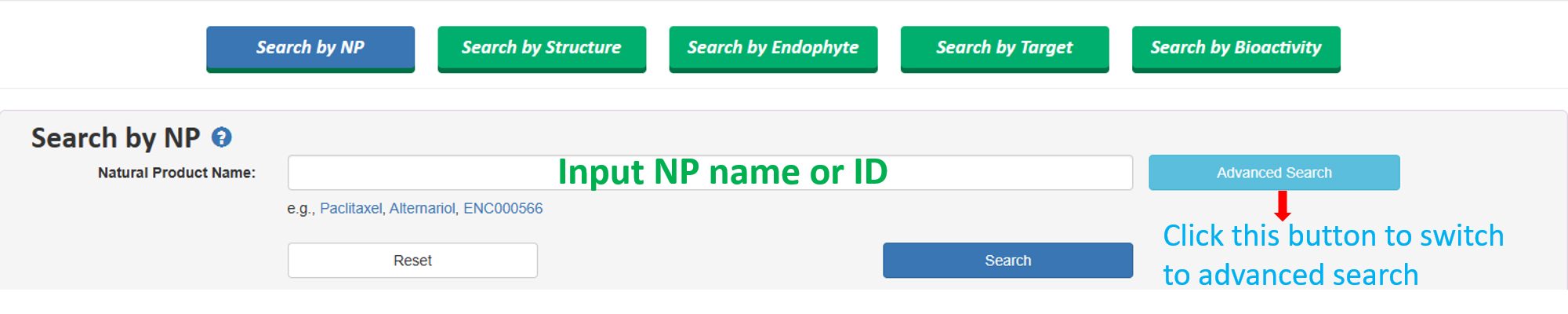
1.1 Search by NPs name/IDs, Advanced search
Users can input compound name (can be common name, synonyms, IUPAC name, or compounnd ID).
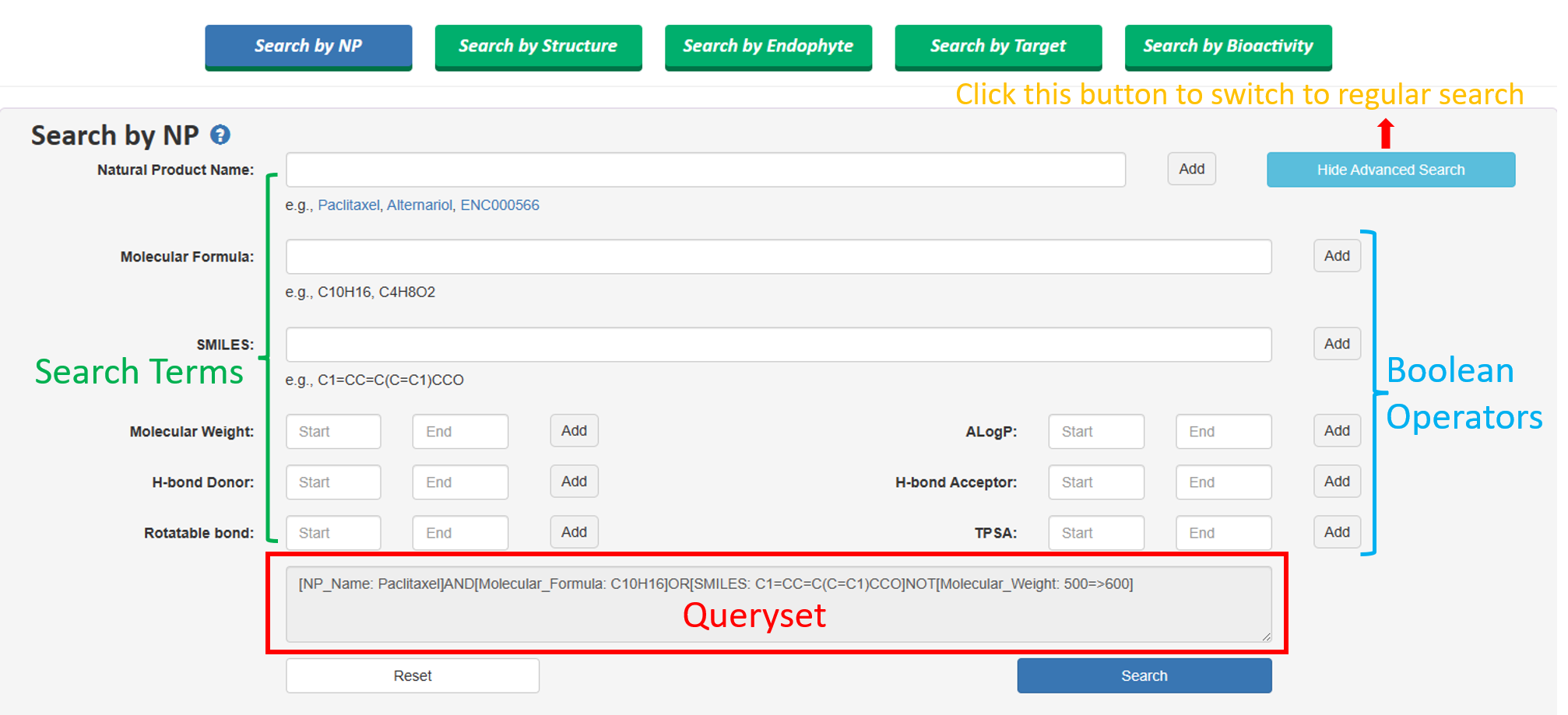
Besides, Users can use one or combination of multiple of these options to customize their search. This allows users to specify any number of query conditions, which can be combined using boolean operators such as "AND," "OR," or "NOT." The available query conditions include molecular formula, SMILES notation, logP range, molecular weight, number of hydrogen bond acceptors (HBA), number of hydrogen bond donors (HBD), number of rotatable bonds, and TPSA range.
1.2 Search by NPs structure
To do a NPs structure search, users can draw a structure and then generate a SMILES string on this page.

Follow the steps below to perform a search:
① Utilize the various tools on the panel to draw the compound structure.
② Click the "Generate SMILES" button to generate the SMILES string of the structure.
③ Adjust the size of the Tanimoto Similarity Threshold by sliding the slider or entering a specific value.
④ Click the "Search" button to search the top 10 NPs based on Tanimoto similarity threshold.
1.3 Search by Endophytic Mircroorganism
Users can search specific endophytic mircroorganism by inputting species name or IDs.
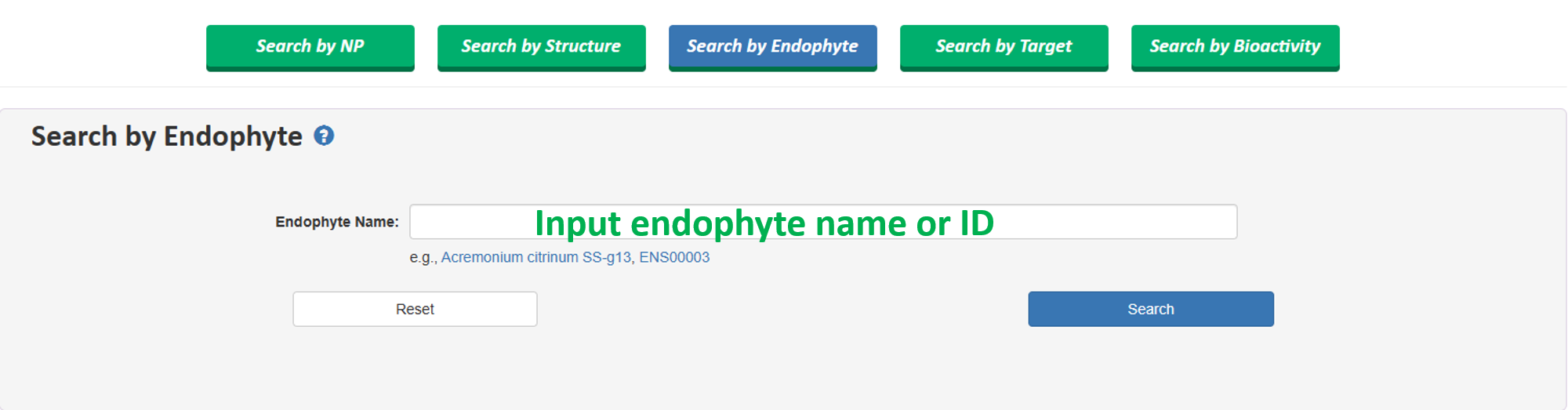
This search supports both exact search and fuzzy search. For example, when searching for the endophytic fungus Acremonium citrinum SS-g13, you can enter the full name 'Acremonium citrinum SS-g13' or simply enter 'Acremonium' to perform the search. Both methods will retrieve 'Acremonium citrinum SS-g13'.
1.4 Search by Target
Users can search a specific target by inputting target name (can be protein name, cell line name and organism name, etc.) or their target IDs.
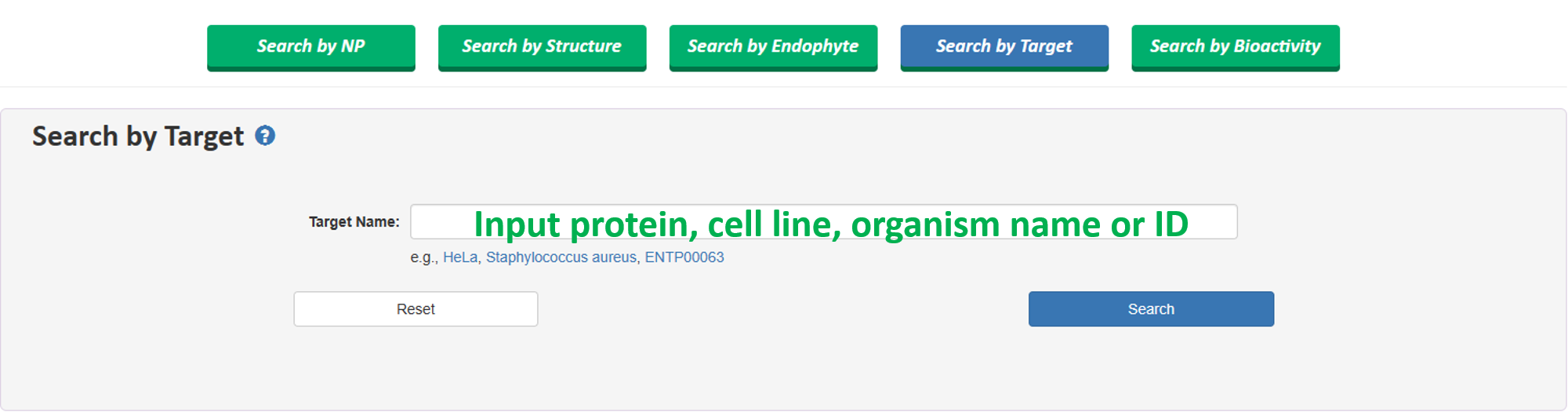
This search supports searching for target name and target ID of proteins, cell lines, and organisms simultaneously. Similarly, this search allows both exact and fuzzy search, and it can retrieve results even with incomplete input by attempting to match as much information as possible.
1.5 Search by Bioactivity
EMNPD offers a dual dropdown component search method for bioactivity. The first dropdown includes bioactivity sizes (Strong, Moderate, Weak, Active, Inactive), and the second dropdown consists of 91 different bioactivities, allowing users to freely combine the contents of these two dropdowns for searching.

2. BROWSE EMNPD
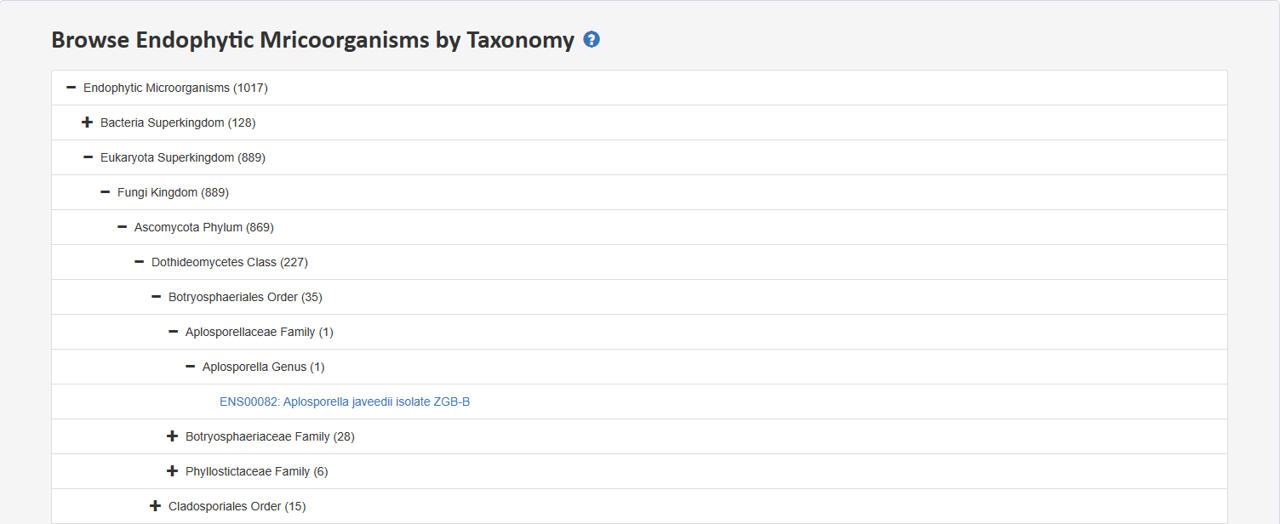
2.1 Browse Endophytic Mricoorganisms by Taxonomy
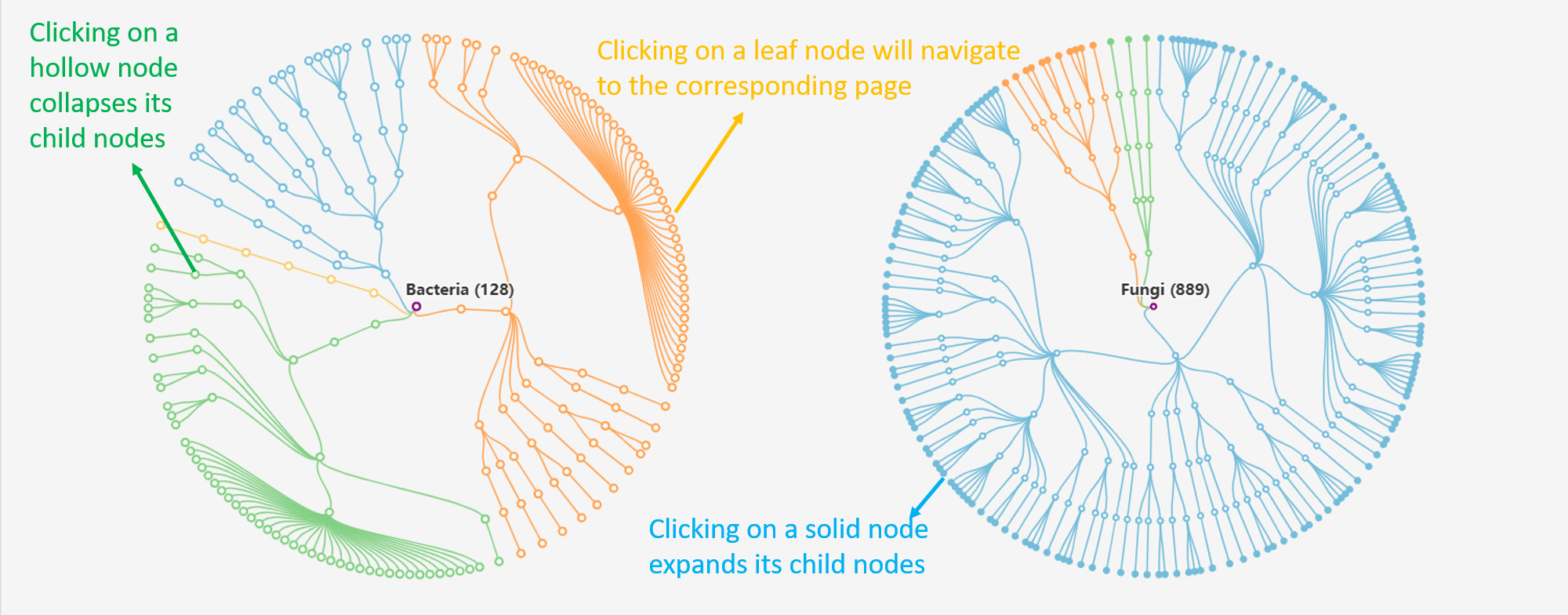
Tree view presents distinct hierarchies of endophyte taxonomy lineage. Users can expand a section of interest to delve deeper into the hierarchy and retrieve specific entries at the last node.
The tree chart collapses the final nodes representing species. Users can expand the solid nodes by clicking on them, allowing access to the corresponding entries.
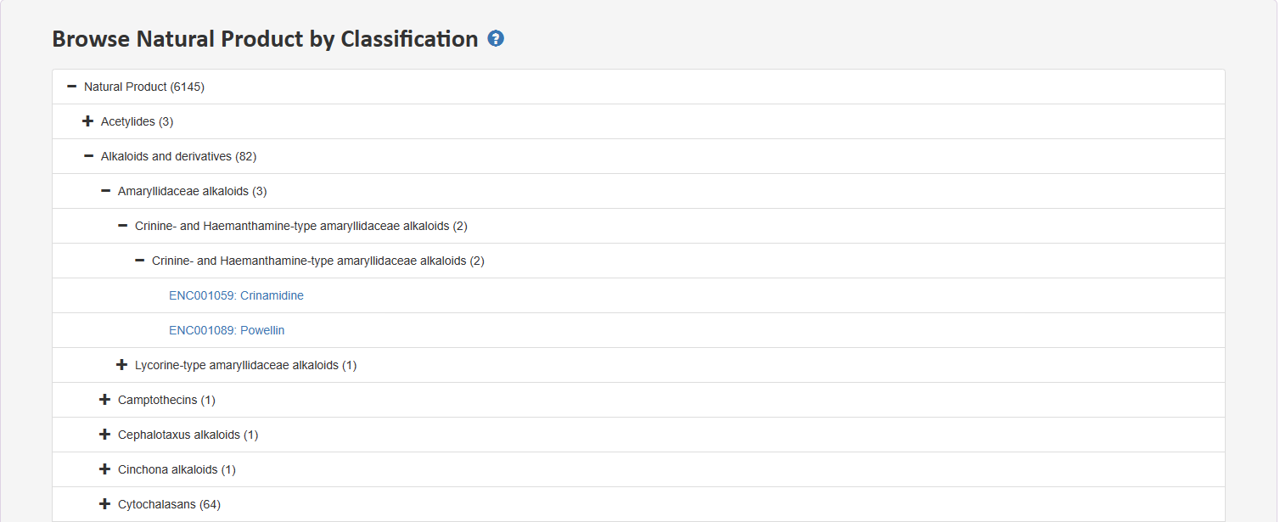
2.2 Browse Natural Product by Classification
Tree view presents distinct hierarchies of NP classification. Users can expand a section of interest to delve deeper into the hierarchy and retrieve specific entries at the last node.
The Bar chart visually represents the distribution ranges of molecular weight, logP, HBA, and HBD for NPs . By clicking on each bar, users can navigate to the corresponding search page with matching criteria.

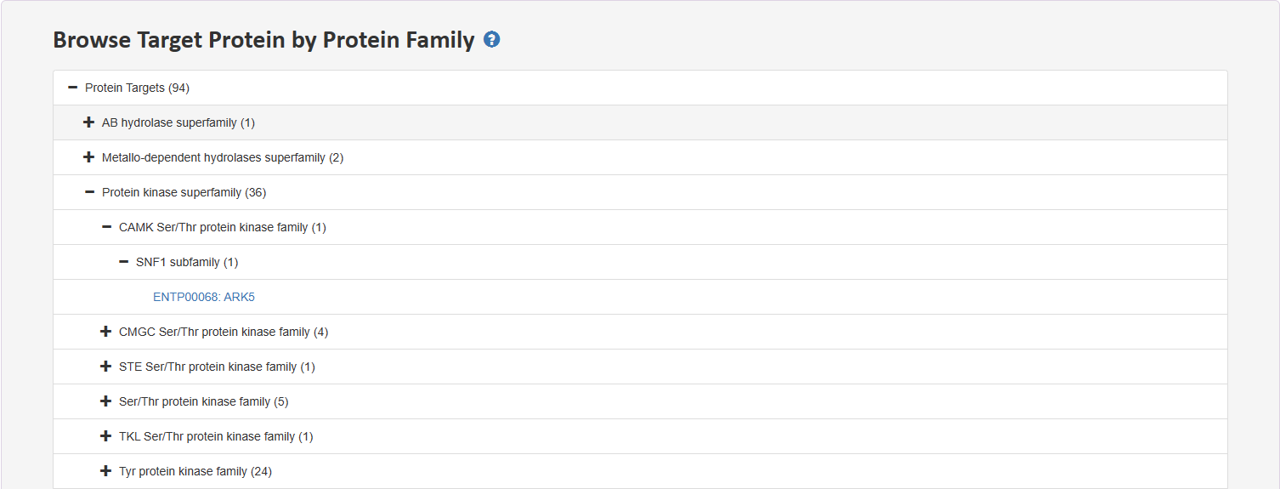
2.3 Browse Target Protein by Protein Family
Tree view presents distinct hierarchies of protein family. Users can expand a section of interest to delve deeper into the hierarchy and retrieve specific entries at the last node.
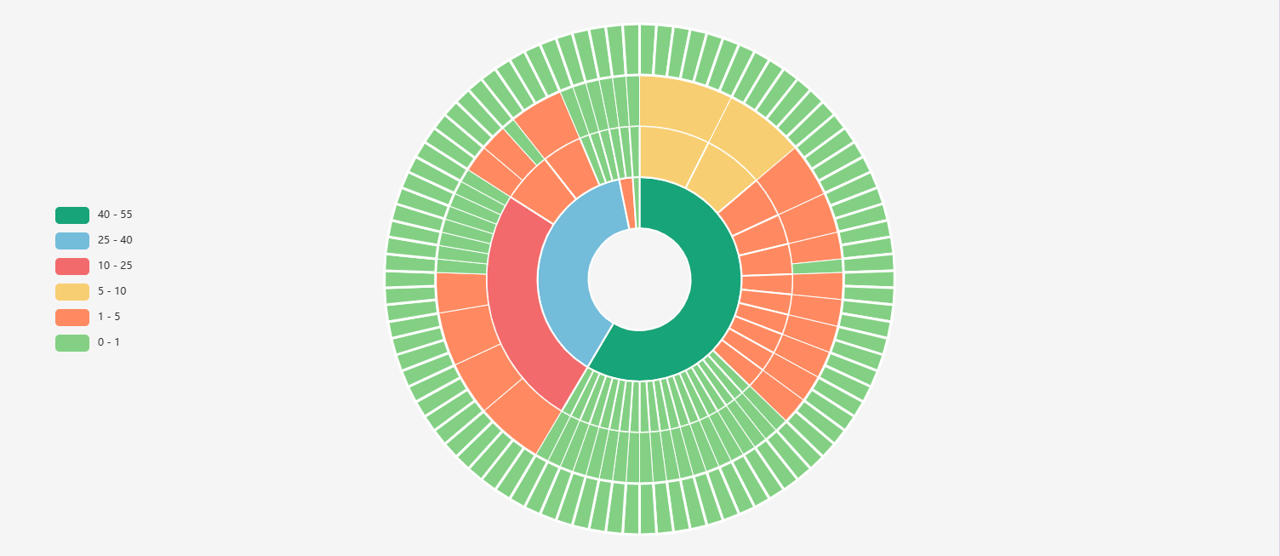
Sunburst chart displays the hierarchical structure of protein family. Representation of the cell line classification hierarchies. Click on a section to expand it and then click on the last node exploring the classification level in more detail.
2.4 Browse Target Cell Line by Category
Tree view presents distinct hierarchies of cell line category. Users can expand a section of interest to delve deeper into the hierarchy and retrieve specific entries at the last node.
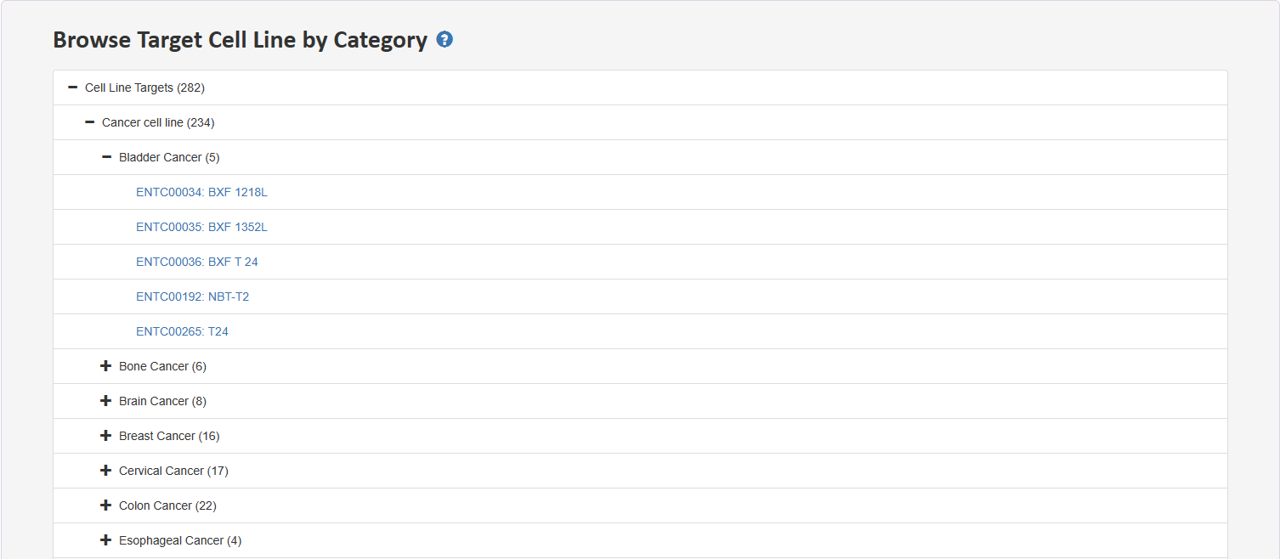
Sunburst chart displays the hierarchical structure of cell line category. Representation of the cell line classification hierarchies. Click on a section to expand it and then click on the last node exploring the classification level in more detail.

2.5 Browse Target Organism by Taxonomy
Tree view presents distinct hierarchies of organism taxonomy lineage. Users can expand a section of interest to delve deeper into the hierarchy and retrieve specific entries at the last node.
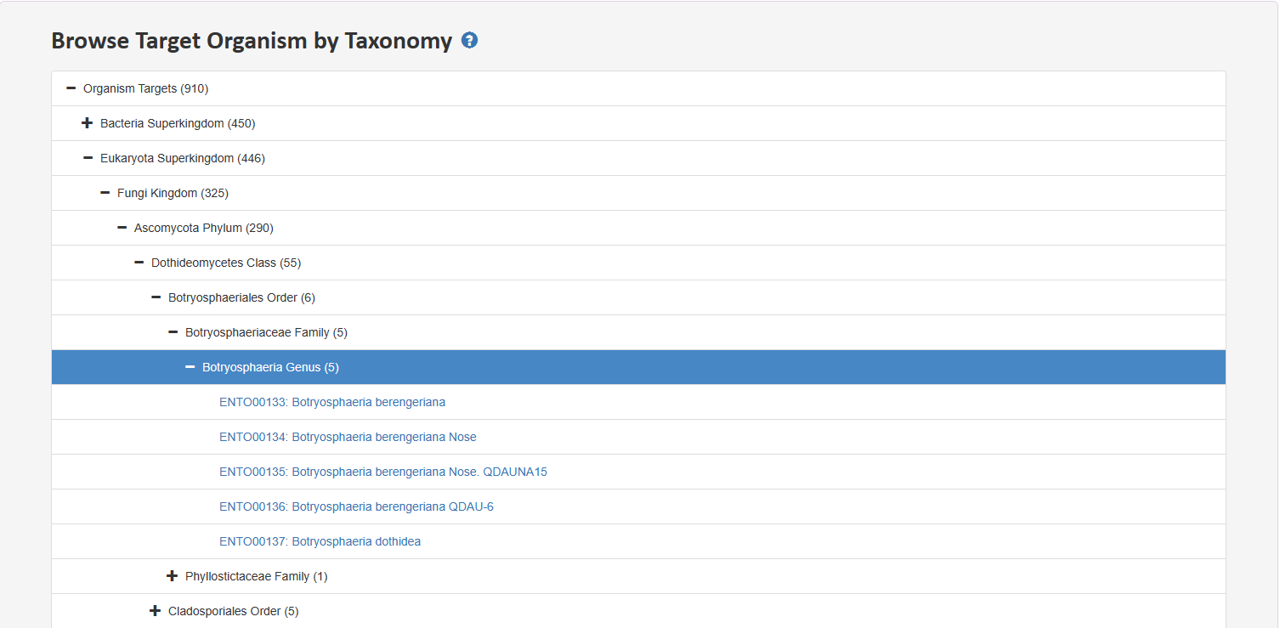
The tree chart collapses the final nodes representing species. Users can expand the solid nodes by clicking on them, allowing access to the corresponding entries.

3. ESCRIPTION OF INDIVIDUAL WEBPAGE
3.1 Webpage of a specific endophytic microorganism
The endophyte page includes three sections: 1. Endophyte Organism Basic Information, 2. Individual Natural Products, 3. NPs Content.
1. Endophyte Organism Basic Information:
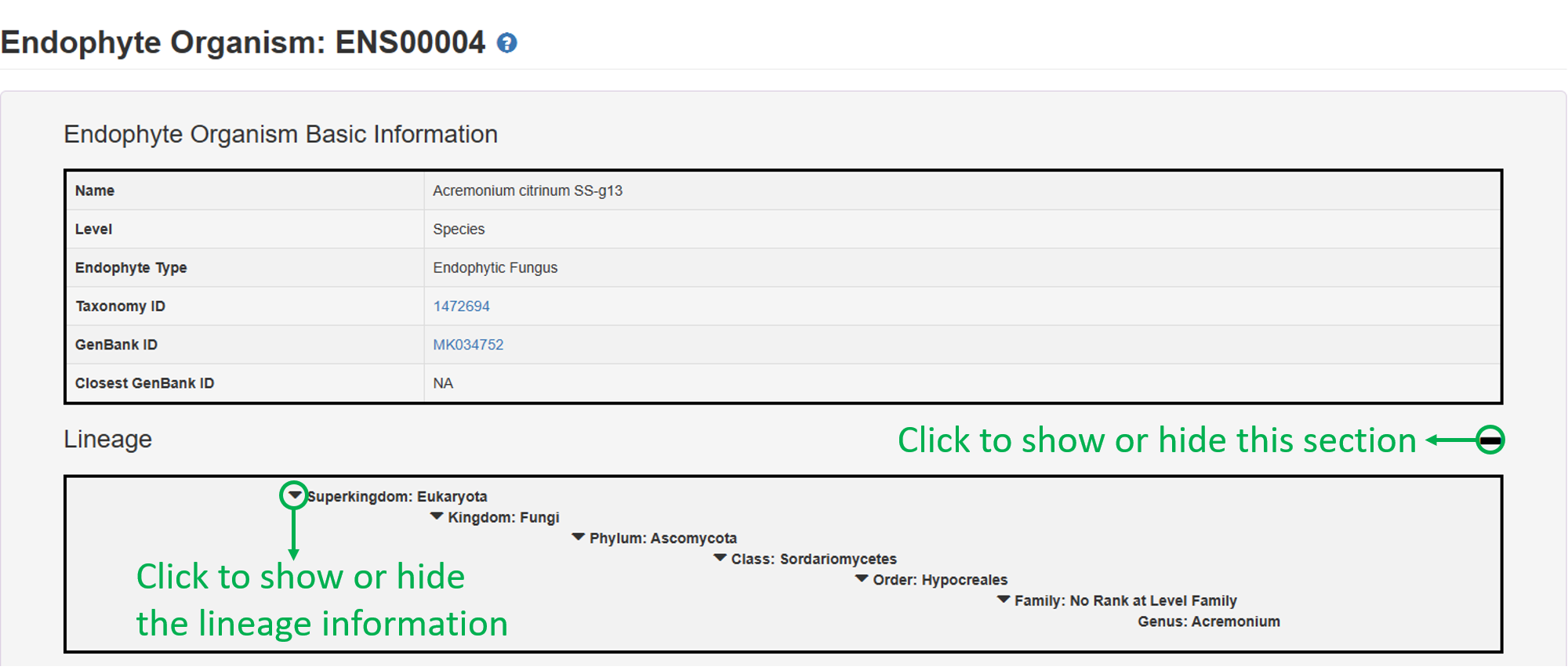
2. Individual Natural Products:

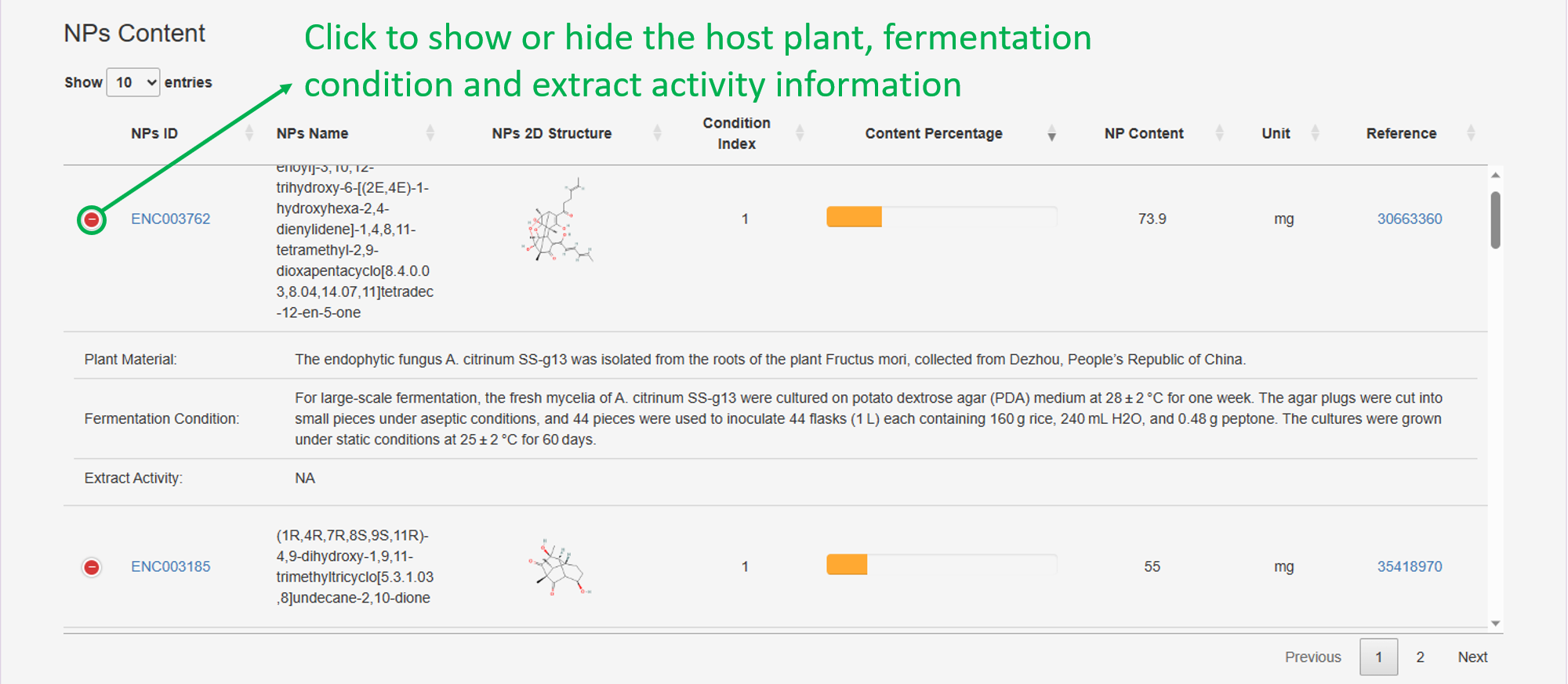
3. NPs Content:
3.2 Webpage of a specific Natural Product
The NP page includes seven sections: 1. NPs Basic Information, 2. NPs Species Source, 3. NPs Content, 4. NPs Biological Activity, 5. NPs Physi-Chem Properties, 6. NPs ADMET Properties, and 7. Similar Compounds.
1. NPs Basic Information:

2. NPs Species Source:

3. NPs Content:

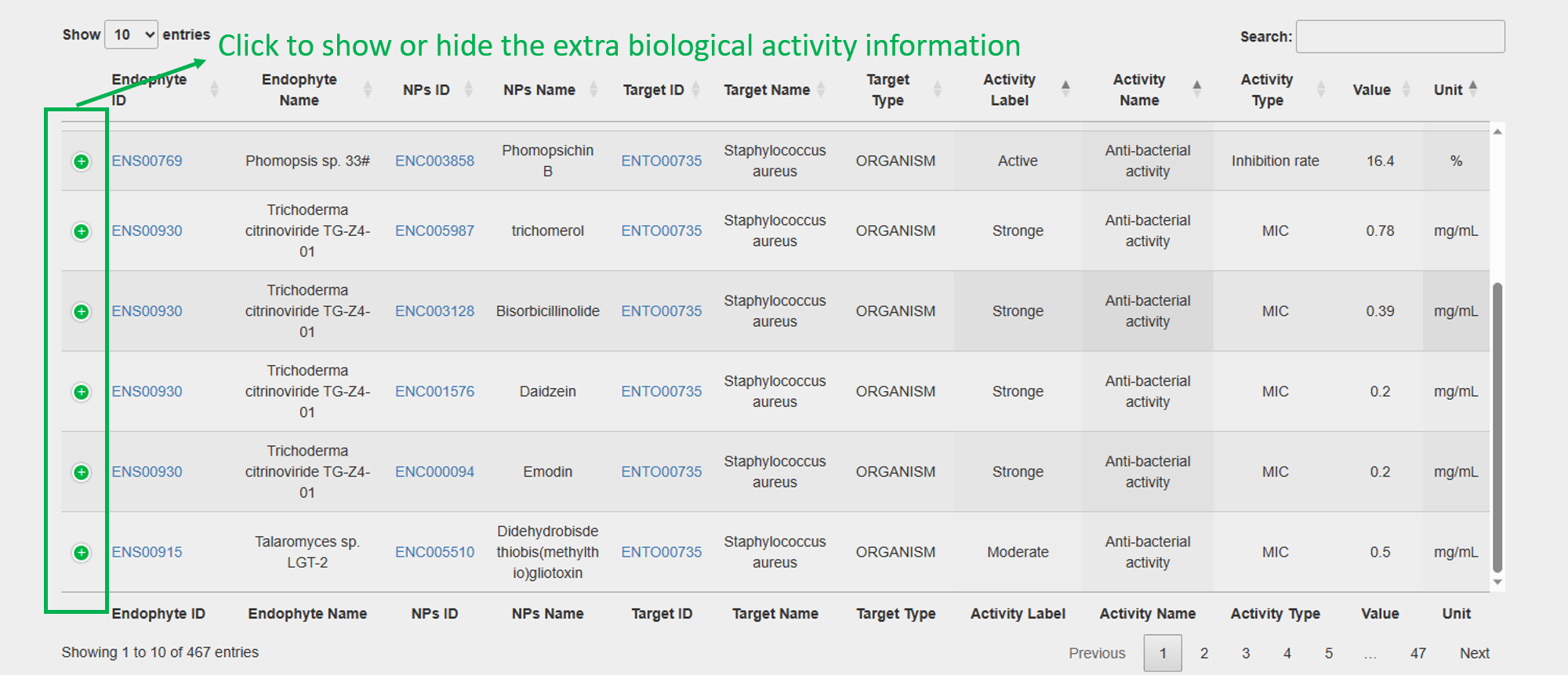
4. NPs Biological Activity:

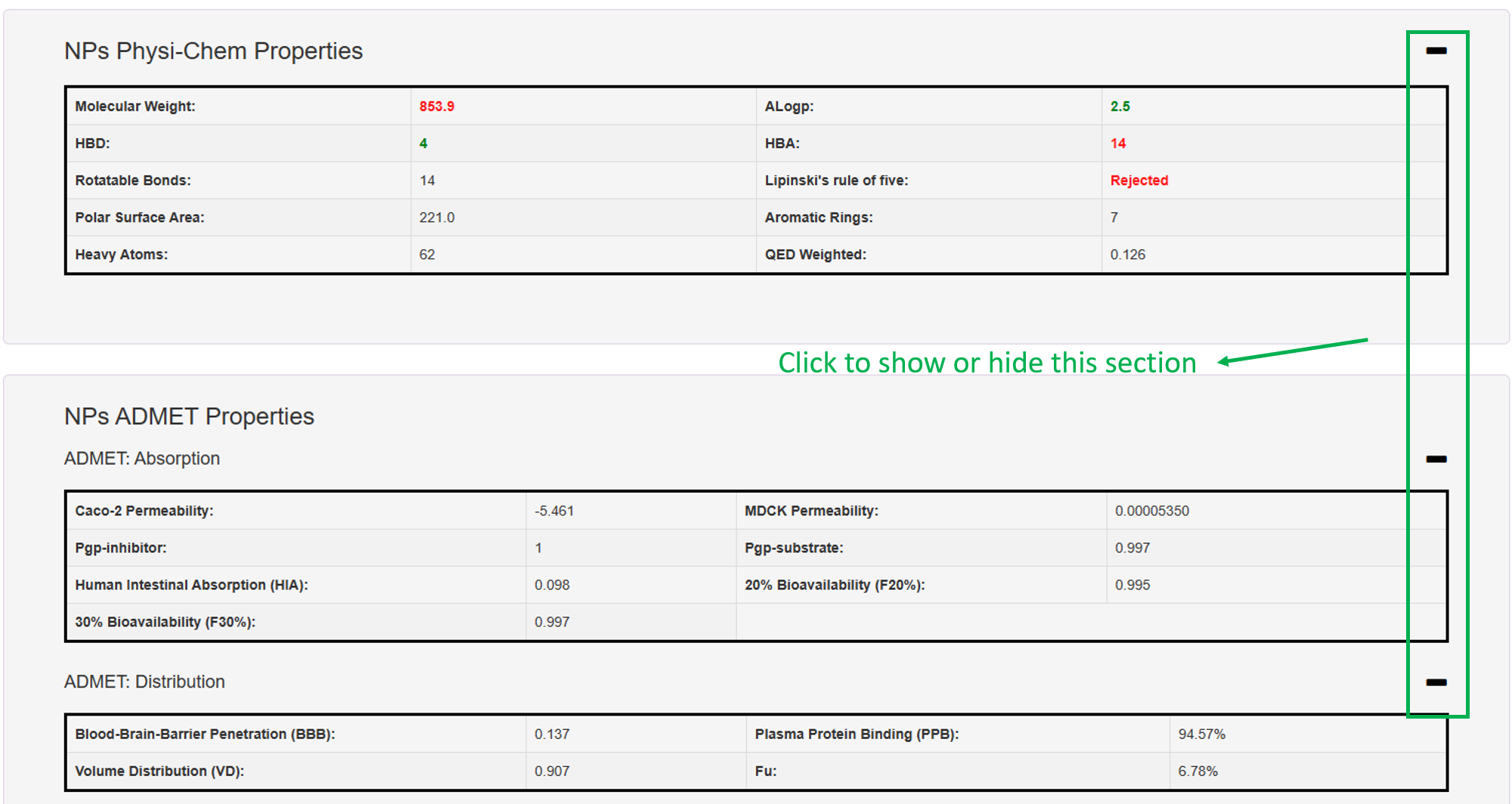
5. NPs Physi-Chem Properties & 6. NPs ADMET Properties:
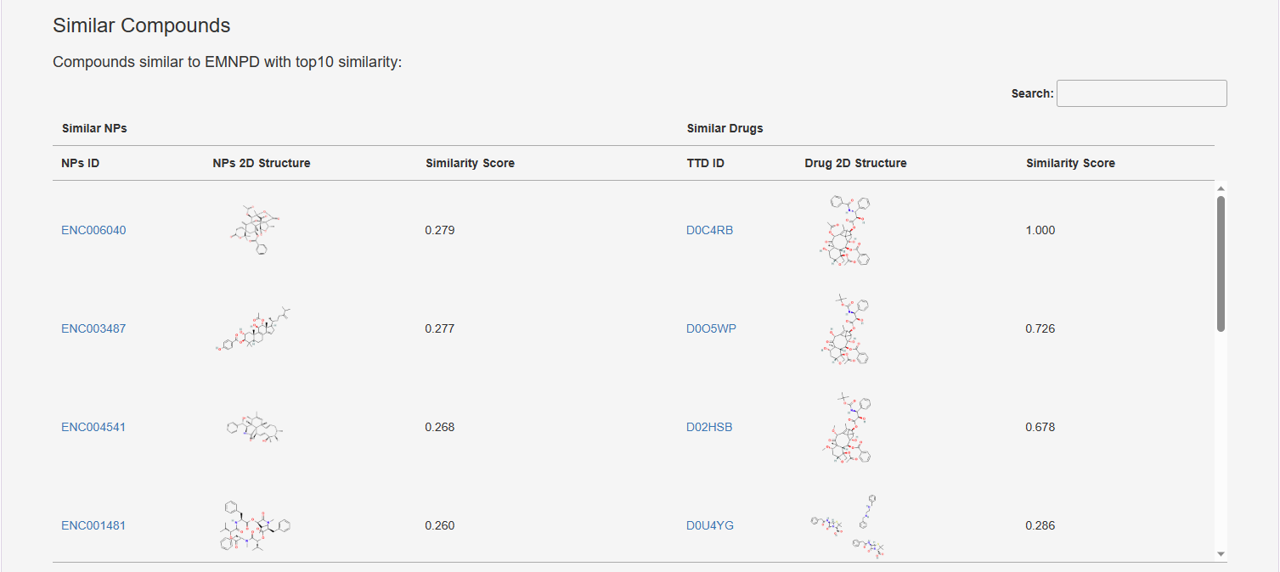
7. Similar Compounds:
3.3 Webpage of a specific target protein
The target protein page includes three sections: 1. Protein Basic Information, 2. Biological Activities of Natural Products against the Target.
1. Protein Basic Information:

2. Biological Activities of Natural Products against the Target:
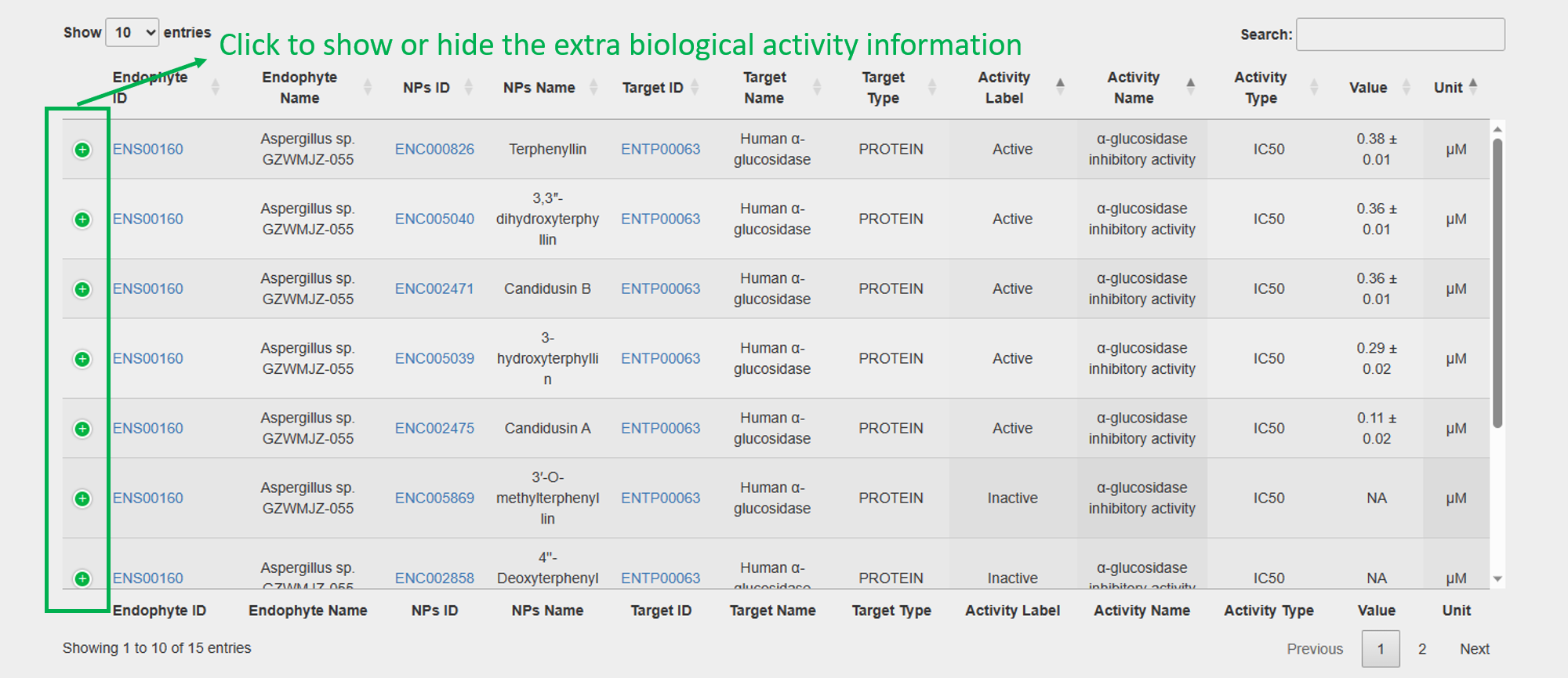
3.4 Webpage of a specific target cell line
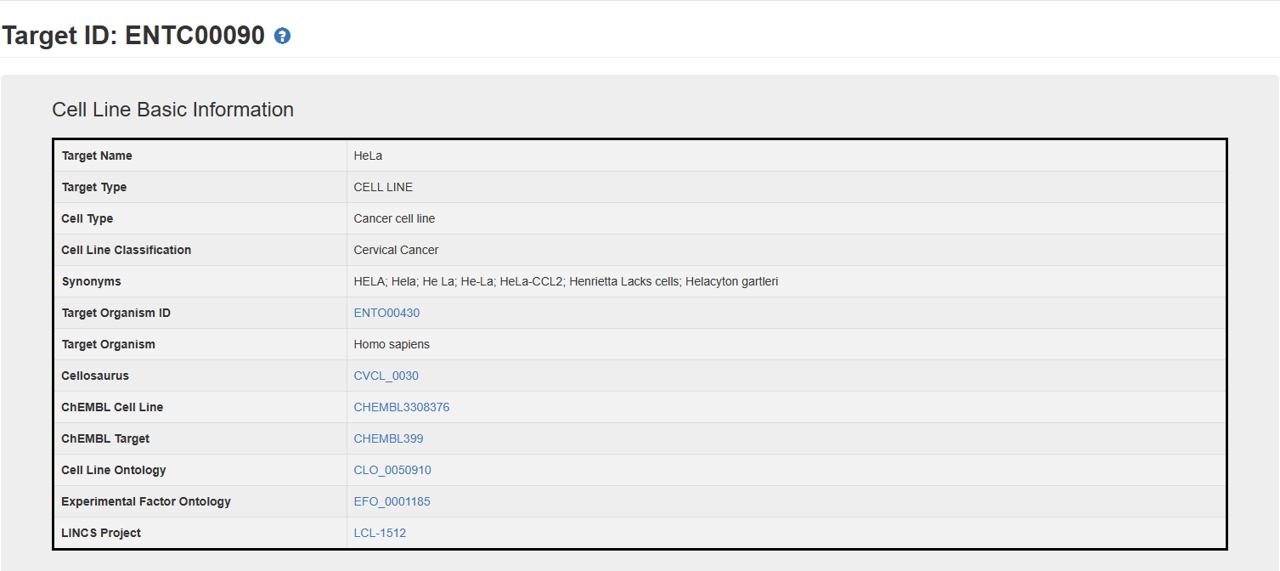
The target cell line page includes three sections: 1. Cell Line Basic Information, 2. Biological Activities of Natural Products against the Target.
1. Cell Line Basic Information:
2. Biological Activities of Natural Products against the Target:

3.5 Webpage of a specific target organism
The target organism page includes three sections: 1. Target Organism Basic Information, 2. Biological Activities of Natural Products against the Target.
1. Target Organism Basic Information:
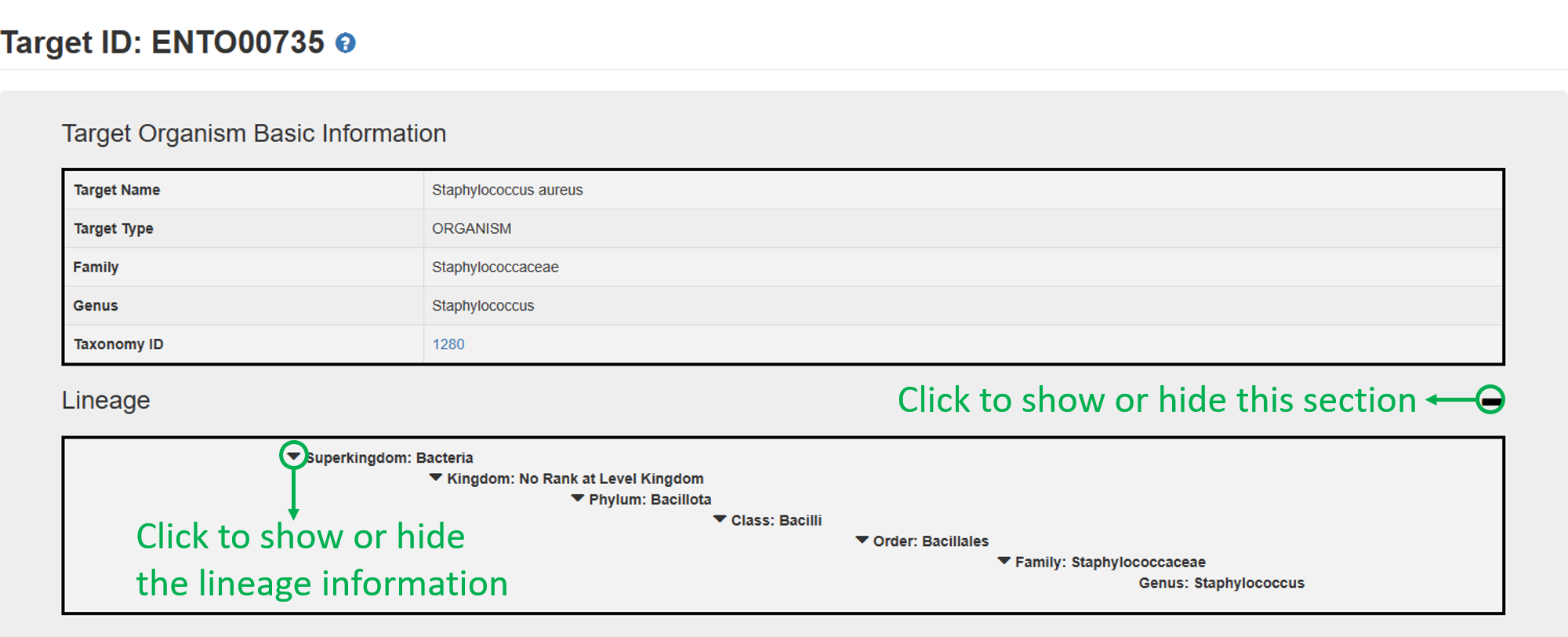
2. Biological Activities of Natural Products against the Target: